Sie befinden sich hier
Inhalt
Background and aim
Recently, the Department of Medicine II established an advanced technology lab to study the dynamics of liver diseases at multiple levels. The required technology was provided by BMBF funding in association to the Systems Biology programs LiSyM and LiSyM-Cancer (2016-2024). The following high-end cutting-edge technologies for in-depth spatial and timely resolved investigations include (1) GeoMX SDP, (2) Slide scanner, (3) Live cell imaging (IncucyteS3), (4) MACS-based cell isolation and the sorting unit, and (5) Applications in preparation (Autostainer; Unit for Multi-photon microscopy and software for AI-based pattern recognition). Although these technologies have been provided to generate data for the BMBF projects LiSyM (recent grant) and C-TIP-HCC (current grant), we broaden the fields of application to other fields in our Department, as well as offer it to cooperation partners within the Core facility platform Mannheim, to exploit the devices´ potential output in the best possible way, now and in the future. We are convinced to offer great forthcoming research opportunities to the UMM by establishing a new core facility “Cell Isolation and Molecular Imaging” in the Department of Medicine II, to centralize and standardize workflow and data robustness.
List of devices in the Department of Medicine II core facility
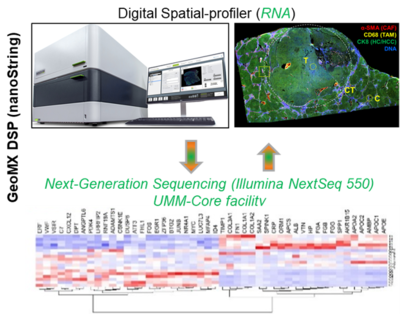
1. GeoMX Digital Spatial Profiler (nanoString)
Spatial biology studies tissues in their natural 2D or 3D context and currently marks the frontier of molecular biology analysis. To this end, we purchased the GeoMX Digital Spatial profiler (DSP; nanoString) by BMBF funds for the C-TIP-HCC project. With the GeoMX DSP technology, we are able to map the spatial architecture of a region of interest (ROI; in this case, cirrhotic tissue with HCC) and visualize how the different liver cell types and cellular phenotypes communicate and interact. Briefly, tissue will be analyzed using formalin-fixed paraffin slides. The tissue will be stained with a multiplex cocktail of labeled antibodies (ab, e.g. for cancer-associated fibroblasts (CAFs), tumour-associated macrophages (TAMs) and epithelial cells), will be probed with barcoded DNA oligos (using whole transcriptome atlas; 18,000 RNA targets), or custom based panels of antibodies. Regions of interest (ROIs) are selected based on ab staining (e.g. alpha SMA, CD68 plus CK8), are selectively detected via UV photocleavable linkers covalently bound to the DNA indexing oligos, and are released by exposure to UV light for collection and deep sequencing, in our case with a powerful sequencer (UMM core facilty, Dr. Tina Fuchs). The obtained data are integrated in a multi-scale model that comprises dynamic, tissue and network models. In addition to liver projects, this technology is currently being used to study for example the treatment response and efficacy in esophageal cancer patients (responder vs. non-responder) upon treatment with immune checkpoint inhibitors (Lancet Healthy Longev 2022; 3: e417–27).
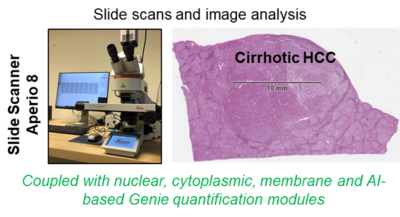
2. Aperio Slide Scanner 8
Aperio Slide Scanner 8 (IHC and IF) coupled with quantification modules (Leica) is a versatile digital pathology scanner with superior image quality and single click operation that is able to highly resolve staining results and greatly facilitate and accelerate spatio-temporal morphological analyses. Having this combined with the ability to digitize slide data at hand, whole slide quantitative measures of e.g. stained mouse and patient samples can be performed immediately upon availability. The core of the running LiSyM-Cancer program (C-TIP-HCC), is the morphological characterization of cirrhotic nodules and HCC by imaging and image analysis. Whole slide scans of human and mouse cirrhotic livers as well as HCC nodules will be generated and analyzed. The process parameters (experimentally suggested as possible TIPping point) includes the number of hepatocyte layers per sheet between neighboring hepatocytes, the contact area (%) of each hepatocyte with the sinusoid, the blood vessel network density & architecture, the bile canalicular architecture, the number of hepatocytes forming canalicular lumen, the hepatocyte nuclear ploidy, the matrisome types and distribution, the vesicle fraction, the NPC densities, the HSC/CAF- and macrophages/TAM-phenotypes, etc. Scans will be quantified and used to generate a tissue-based mathematical model. Later, this data will be used to predict/validate a TIP in cirrhotic patients. In addition to liver studies, we are convinced that the available slide scanner (combined with image analysis modules) can and will be used to generate high-quality images and quantification opportunities for multiple projects of our Department and can also be applied to other collaborative projects of the faculty.
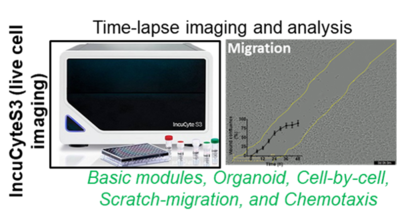
3. IncuCyte S3 Live-Cell Analysis System coupled with quantification modules (Sartorius)
The Live-cell imaging platform offers time-lapse studies of kinetic processes such as, migration, chemoresistence, apoptosis, proliferation chemotaxis, enzyme activity and signal transduction to study cellular fate and cell-cell interaction, high throughput and walk-away in a full-automated live cell imaging manner. Using extra-funding from the LiSyM consortium (2016-2021), the live cell imaging-IncuCyteS3 platform was established. Additionally, with BMBF funding for the subsequent LiSyM-Cancer (C-TIP-HCC) consortium (2021-2024), the InuCyte modules for the analysis of organoids, cell migration, cell subpopulation phenotyping, and chemotaxis have been purchased. Data thus obtained will contribute significantly to the achievement of various C-TIP-HCC milestones and other DFG funded project of our AG. This live cell imaging platform offers diverse research opportunities for other projects at the Department of Medicine II. (i.e. Organoid-related research topics of PD Dr. Johannes Betge and PD Dr. Elke Burgermeister) and again for other interested collaborators at the UMM.
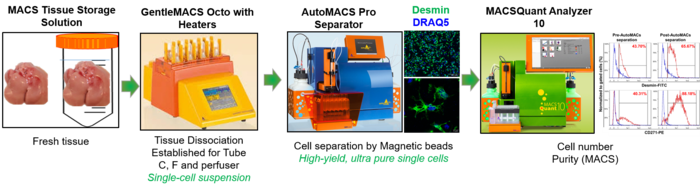
4. AutoMACS Pro Separator-cell isolation coupled with MACSQuant and tissue disscoiator (miltenyi Biotec) Cell Purification Unit (CPU)
Basic researchers using the current SOP for isolation of primary cells face several limitations, i.e. time-consuming and sensitive steps. Further, automatization was one important step to increase reproducibility. Therefore, we established a unit for fully automated cell isolation, specifically from fresh liver tissue as well as from storage solution preserved tissue upon shipment, for improved and more robust and reproducible downstream investigations, including RNA and protein analyses. This SOP is based on a fully-automated cell labelling and separation procedure using Miltenyi Biotec technologies (Tissue dissociator, autoMACS Separator and MACSQuant). Besides using this unit to isolate primary liver cells, we also offer this technology for the entire faculty.
List of publications with support from Cell Isolation and Molcular Imaging facility
- Chen Y, Lin J, Schlotterer A, Kurowski L, Hoffmann S, Hammad S, Dooley S, Buchholz M, Hu J, Fleming I, Hammes HP. MicroRNA-124 Alleviates Retinal Vasoregression via Regulating Microglial Polarization. Int J Mol Sci. 2021 Oct 14;22(20):11068. PMID: 34681723
- Colucci S, Altamura S, Marques O, Dropmann A, Horvat NK, Müdder K, Hammad S, Dooley S, Muckenthaler MU. Liver Sinusoidal Endothelial Cells Suppress Bone Morphogenetic Protein 2 Production in Response to TGFβ Pathway Activation. Hepatology. 2021 Oct;74(4):2186-2200. PMID: 33982327
- Hammad S, Ogris C, Othman A, Erdoesi P, Schmidt-Heck W, Biermayer I, Helm B, Gao Y, Piorońska W, Holland CH, D'Alessandro LA, de la Torre C, Sticht C, Al Aoua S, Theis FJ, Bantel H, Ebert MP, Klingmüller U, Hengstler JG, Dooley S, Mueller NS. Tolerance of repeated toxic injuries of murine livers is associated with steatosis and inflammation. Cell Death Dis. 2023 (Provisionally accepted)
- Hoehme S, Hammad S, Boettger J, Begher-Tibbe B, Bucur P, Vibert E, Gebhardt R, Hengstler JG, Drasdo D. Digital twin demonstrates significance of biomechanical growth control in liver regeneration after partial hepatectomy. iScience. 2022 Dec 5;26(1):105714. PMID: 36691615
- Manta CP, Leibing T, Friedrich M, Nolte H, Adrian M, Schledzewski K, Krzistetzko J, Kirkamm C, David Schmid C, Xi Y, Stojanovic A, Tonack S, de la Torre C, Hammad S, Offermanns S, Krüger M, Cerwenka A, Platten M, Goerdt S, Géraud C. Targeting of Scavenger Receptors Stabilin-1 and Stabilin-2 Ameliorates Atherosclerosis by a Plasma Proteome Switch Mediating Monocyte/Macrophage Suppression. Circulation. 2022 Dec 6;146(23):1783-1799. PMID: 36325910
Kontextspalte
Ansprechpartner

Das Forschungsnetzwerk Leber-Systemmedizin LiSyM ist ein Netzwerk von 20 deutschen Forschungszentren. Vom Bundesministerium für Bildung und Forschung wird der Verbund mit 20 Millionen Euro für die Dauer von 5 Jahren unterstützt.